1. β-Hydroxybutyrate suppresses colorectal cancer | Nature
Here we identify a metabolite signalling pathway that provides actionable insights towards this goal.
We perform a dietary screen in autochthonous animal models of CRC and find that ketogenic diets exhibit a strong tumour-inhibitory effect.
These properties of ketogenic diets are recapitulated by the ketone body β-hydroxybutyrate (BHB), which reduces the proliferation of colonic crypt cells and potently suppresses intestinal tumour growth.
We find that BHB acts through the surface receptor Hcar2 and induces the transcriptional regulator Hopx, thereby altering gene expression and inhibiting cell proliferation.
Cancer organoid assays and single-cell RNA sequencing of biopsies from patients with CRC provide evidence that elevated BHB levels and active HOPX are associated with reduced intestinal epithelial proliferation in humans. This study thus identifies a BHB-triggered pathway regulating intestinal tumorigenesis and indicates that oral or systemic interventions with a single metabolite may complement current prevention and treatment strategies for CRC.
2. Somatic genomic changes in single Alzheimer’s disease neurons | Nature
나이가 들면서 뉴런들은 somatic mutation 들을 계속 모으게 되는데, 유전적 요소, 환경에 대한 노출이 원인이 될 수 있음. 이 논문에서는 319 개 뉴런에 scWGS를 돌려서 나온 결과를 봤음. prefrontal cortex and hippocampus of individuals with Alzheimer’s disease.
Mutational signature 분석 결과 일반 뉴런에서는 age-related pattern 이 주로 발견되었고, 알츠하이머 샘플에서는 Signature C 가 발견되었는데 이는 nucleotide oxidation 에 의해 일어날 수 있음. ( In neurons affected by Alzheimer’s disease, additional DNA alterations are driven by distinct processes (signature C) that highlight C>A and other specific nucleotide changes. These changes potentially implicate nucleotide oxidation4,11, which we show is increased in Alzheimer’s-disease-affected neurons in situ.)
발현된 gene 들도 signature-specific damage 를 나타내고, 변이들도 transcriptional strand bias 를 나타내는데 이는 transcription-coupled nucleotide excision repair 가 mutation 의 원인이 되는데 역할을 했음을 보여줌.
The alterations in Alzheimer’s disease affect coding exons and are predicted to create dysfunctional genetic knockout cells and proteostatic stress.
3. Stepwise-edited, human melanoma models reveal mutations’ effect on tumor and microenvironment (science.org)

여기서도 여러가지 mutation 을 한꺼번에 봐야 하는 중요한 이유를 설명하는데, 이는 cancer 의 특성 상 하나의 mutation 이 drive 하는 경우가 많지 않다는 것. 결국에는 여러 mutation 에 대한 systematic 분석이 필요하다는 것이고, 여기서는 여러 mutation 들을 genome edit 을 통해 만들어가며 cell -> mouse model -> in-vitro human application 까지 가는 것을 보여주고 있음.
Normal cell -> induced specific cell state -> to mouse cancer model -> sc analysis
Mouse cancer model -> tumor isolation -> tumor cell decomposition -> stromal cell -> tumor microenvironment analysis
Mouse cancer model -> tumor isolation-> tumor histopathological image -> Deep learning modeling (included pre-trained top-tier image classification model) -> Tested on real-life human sample
로 이어지는 논문.
4. Cancer cells use self-inflicted DNA breaks to evade growth limits imposed by genotoxic stress (science.org)
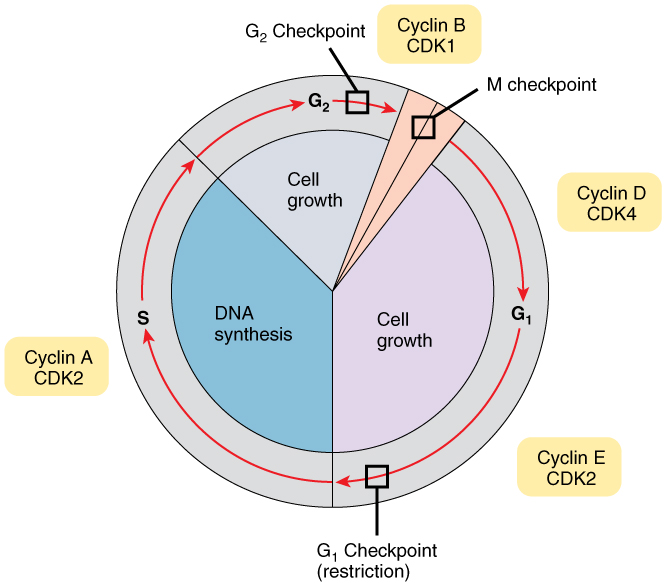
일반 세포들은 genotoxic insult ( DNA damage 등 ) 을 받으면 G1 cell cycle checkpoint 를 활성화시켜서 세포 성장을 막는데, 종양세포에서는 이것이 작동을 안한다.
Larsen et al. 에서는 종양세포가 nuclease 를 활성화시켜서 특정 부위에 DNA break 를 만든 다음에, DNA break repair 를 유도한다는 연구결과를 냈는데, 이 DNA 조각들은 G2 cell cycle checkpoint 를 작동시켜서, treatment 후에 DNA 가 망가져도 ( tumor cell 이 공격을 받아도 ) 죽지 않고 살게 한다고 한다.
caspase-activated DNase ( CAD ) 에 의해서 일어나고, CTCF 가까이에 있는 loci 에서 생긴다. CAD 는 인산화 과정에 의해서 조절된다.
5. Half a decade of graph convolutional networks | Nature Machine Intelligence
( Full text 요청중 )
6. RNA folding using quantum computers (plos.org)
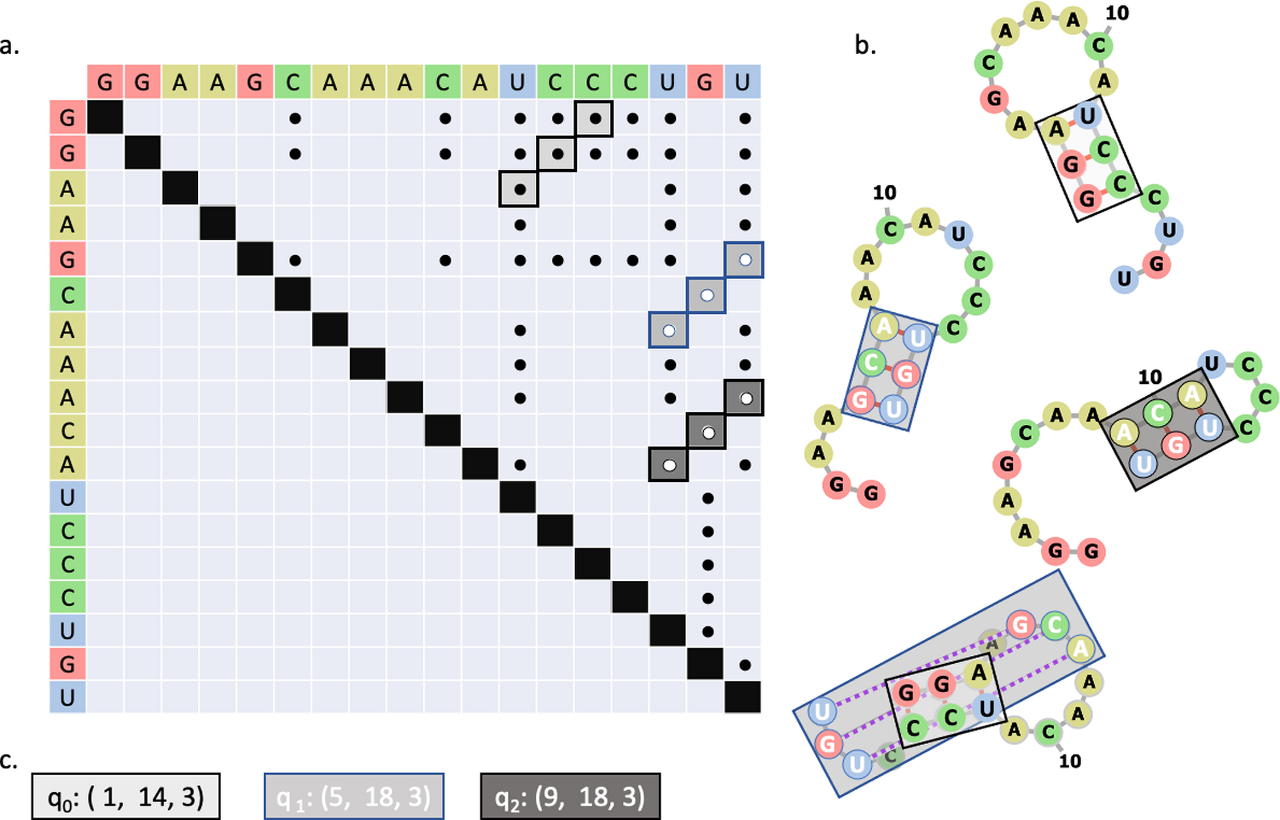
The 3-dimensional fold of an RNA molecule is largely determined by patterns of intramolecular hydrogen bonds between bases.
Predicting the base pairing network from the sequence, also referred to as RNA secondary structure prediction or RNA folding, is a nondeterministic polynomial-time (NP)-complete computational problem.
The structure of the molecule is strongly predictive of its functions and biochemical properties, and therefore the ability to accurately predict the structure is a crucial tool for biochemists. Many methods have been proposed to efficiently sample possible secondary structure patterns.
Classic approaches employ dynamic programming, and recent studies have explored approaches inspired by evolutionary and machine learning algorithms.
This work demonstrates leveraging quantum computing hardware to predict the secondary structure of RNA.
A Hamiltonian written in the form of a Binary Quadratic Model (BQM) is derived to drive the system toward maximizing the number of consecutive base pairs while jointly maximizing the average length of the stems.
A Quantum Annealer (QA) is compared to a Replica Exchange Monte Carlo (REMC) algorithm programmed with the same objective function, with the QA being shown to be highly competitive at rapidly identifying low energy solutions.
The method proposed in this study was compared to three algorithms from literature and, despite its simplicity, was found to be competitive on a test set containing known structures with pseudoknots.
7. Distinguishing excess mutations and increased cell death based on variant allele frequencies (plos.org)
Tumors often harbor orders of magnitude more mutations than healthy tissues. The increased number of mutations may be due to an elevated mutation rate or frequent cell death and correspondingly rapid cell turnover, or a combination of the two.
It is difficult to disentangle these two mechanisms based on widely available bulk sequencing data, where sequences from individual cells are intermixed and, thus, the cell lineage tree of the tumor cannot be resolved.
Here we present a method that can simultaneously estimate the cell turnover rate and the rate of mutations from bulk sequencing data.
Our method works by simulating tumor growth and finding the parameters with which the observed data can be reproduced with maximum likelihood. Applying this method to a real tumor sample, we find that both the mutation rate and the frequency of death may be high.
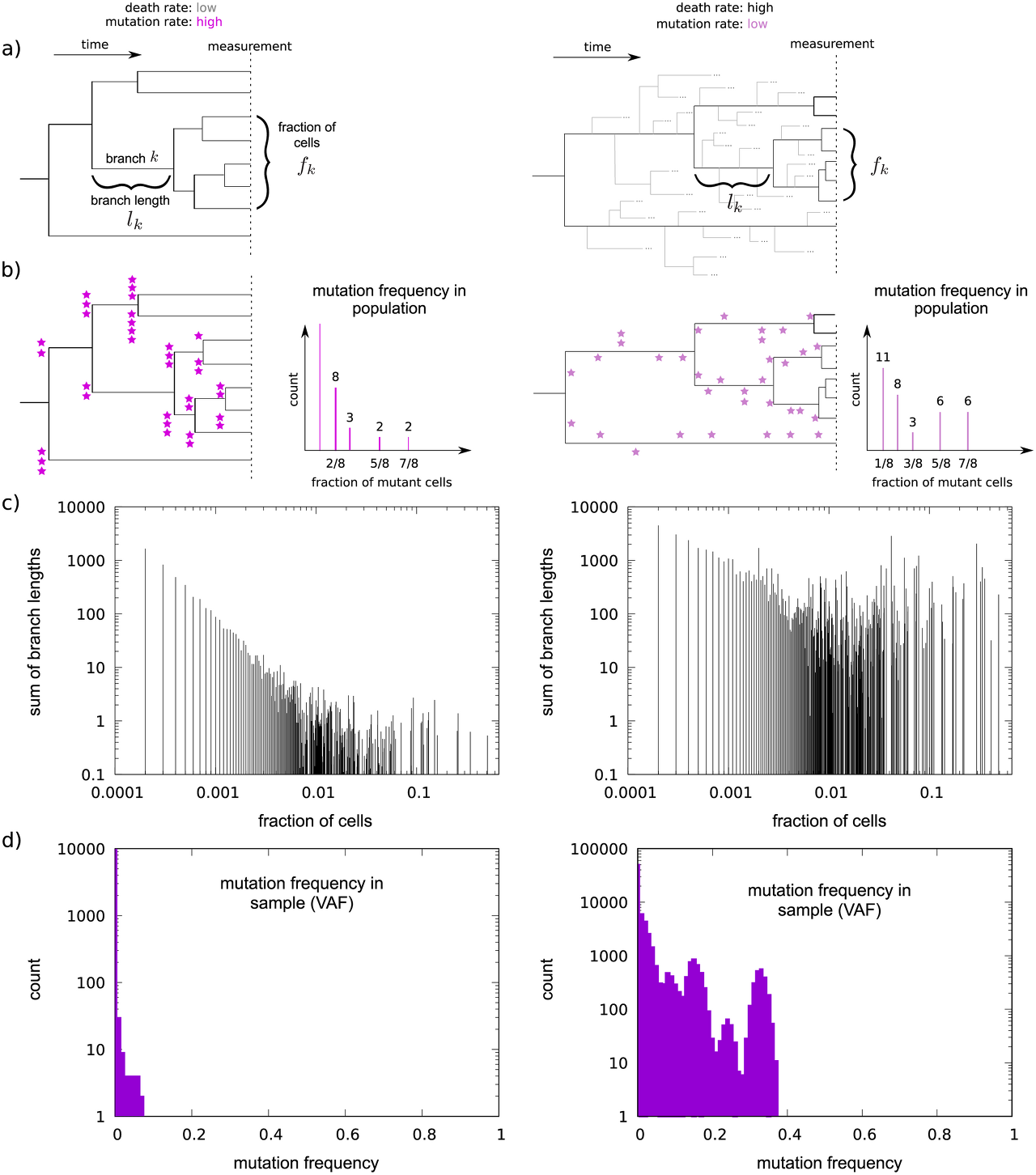
Two possible scenarios for the generation of mutations along cell lineage trees.
A) Different death-to-birth ratios lead to different lineage tree shapes. Bifurcations are cell divisions, leaves are cells comprising the bulk sequencing sample. Note that the (surviving) tree topologies are the same, only branch lengths differ.
B) Mutations, symbolized by purple stars, accumulate at cell divisions. High death-to-birth ratio and low mutation rate can lead to the same number of observed mutations as low death-to-birth ratio and high mutation rate, however, the mutation spectrum of the two trees are different.
C) For simulated trees of 104 leaves, the differences in the branch length distribution are clearly visible.
D) VAF spectra for death-to-birth ratios of 0 (left panel) and 0.999999 (right panel). See also Fig A in S1 Text, which shows the effect of varying the mutation rate on the VAF spectra. The mutation rate was set to μ = 1. Fractions of mutant cells are binned (note the logarithmic scale). Ploidy is set to two, contamination is zero. Simulated sequencing depth is 1000. The VAF spectra are based on the trees used to generate subplot C).
https://doi.org/10.1371/journal.pcbi.1010048.g001
8. Inferring ongoing cancer evolution from single tumour biopsies using synthetic supervised learning (plos.org)
Inferring ongoing cancer evolution from single tumour biopsies using synthetic supervised learning
Author summary Recent pioneering work modeling the evolutionary dynamics in growing tumours has provided fundamental insight into the quantitative relationship between mutation frequency and evolutionary parameters. However, existing approaches to infer ca...
journals.plos.org
Variant allele frequencies (VAF) encode ongoing evolution and subclonal selection in growing tumours. However, existing methods that utilize VAF information for cancer evolutionary inference are compressive, slow, or incorrectly specify the underlying cancer evolutionary dynamics.
Here, we provide a proof-of-principle synthetic supervised learning method, TumE, that integrates simulated models of cancer evolution with Bayesian neural networks, to infer ongoing selection in bulk-sequenced single tumour biopsies. Analyses in synthetic and patient tumours show that TumE significantly improves both accuracy and inference time per sample when detecting positive selection, deconvoluting selected subclonal populations, and estimating subclone frequency. Importantly, we show how transfer learning can leverage stored knowledge within TumE models for related evolutionary inference tasks—substantially reducing data and computational time for further model development and providing a library of recyclable deep learning models for the cancer evolution community. This extensible framework provides a foundation and future directions for harnessing progressive computational methods for the benefit of cancer genomics and, in turn, the cancer patient.
9. The future of early cancer detection | Nature Medicine
( Full text 요청중 )


댓글